Introduction to Pydicom
December 31, 2020
Code can be found @ Github. Checkout dicom-viewer.ipynb
Abstract
Pydicom is a pure Python package for working with DICOM files. It lets you read, modify and write DICOM data in an easy “pythonic” way.
Introduction
Installation
Using pip:
$ pip install pydicom
Using conda:
$ conda install -c conda-forge pydicom
Pydicom comes with its own set of dicom images which can be used to go through examples.
They also give get_dataset.py file to download datasets, which is also included in the github repo.
$ python get_datasets.py --show
$ python get_datasets.py --output {path}
Tutorial
def load_scan(path):
slices = [pydicom.dcmread(path + '/' + s) for s in
os.listdir(path)]
slices = [s for s in slices if 'SliceLocation' in s]
slices.sort(key = lambda x: int(x.InstanceNumber))
try:
slice_thickness = np.abs(slices[0].ImagePositionPatient[2] -
slices[1].ImagePositionPatient[2])
except:
slice_thickness = np.abs(slices[0].SliceLocation -
slices[1].SliceLocation)
for s in slices:
s.SliceThickness = slice_thickness
return slices
load_scan loads the dicom files and sorts them according to their Instance Number. It also extracts slice thickness which is a very important parameter in these scans as it is indicative of resolution of the scans. Lower the slice thickness, better the resolution.
Slice Thickness
Slice thickness directly impacts the precision of target localization during treatment.
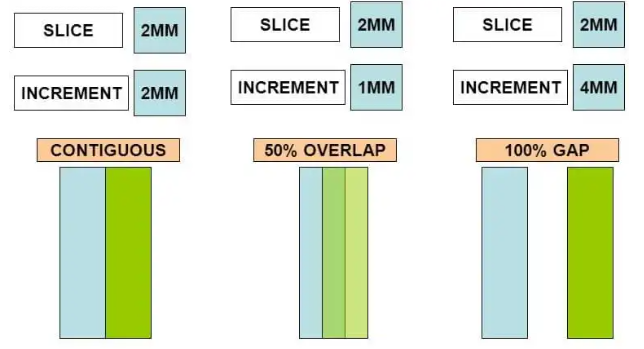
Slice Increment/Spacing refers to the movement of the table/scanner for scanning the next slice.
If slice thickness is greater than slice increment than there is anatomical information loss.
If there is overlap between 2 adjacent slices that is slice thickness > slice increment than such cases acts as error correction.
HU Scaling
def get_pixels_hu(scans):
image = np.stack([s.pixel_array for s in scans])
image = image.astype(np.int16)
# Convert to Hounsfield units (HU)
slope = scans[0].RescaleSlope
if slope != 1:
image = slope * image.astype(np.float64)
image = image.astype(np.int16)
image += np.int16(scans[0].RescaleIntercept)
return np.array(image, dtype=np.int16)
HU scaling is explained in my dicom standard blog.
Multiplanar reconstruction
slices = sorted(patient_dicom, key=lambda s: s.SliceLocation)
# pixel aspects, assuming all slices are the same
pixel_spacing = slices[0].PixelSpacing
slice_thickness = slices[0].SliceThickness
ax_aspect = pixel_spacing[1]/pixel_spacing[0]
sag_aspect = pixel_spacing[1]/slice_thickness
cor_aspect = slice_thickness/pixel_spacing[0]
# create 3D array
img_shape = list(slices[0].pixel_array.shape)
img_shape.append(len(slices))
img3d = np.zeros(img_shape)
# fill 3D array with the images from the files
for i, s in enumerate(slices):
img2d = s.pixel_array
img3d[:, :, i] = img2d
# print(img3d.shape)
# plot 3 orthogonal slices
a1 = plt.subplot(1, 3, 1)
a1.axis('off')
plt.imshow(img3d[:, :, img_shape[2]//2], cmap='gray')
a1.set_aspect(ax_aspect)
a2 = plt.subplot(1, 3, 2)
a2.axis('off')
plt.imshow(img3d[:, img_shape[1]//2, :], cmap='gray')
a2.set_aspect(sag_aspect)
a3 = plt.subplot(1, 3, 3)
a3.axis('off')
plt.imshow(img3d[img_shape[0]//2, :, :].T, cmap='gray')
a3.set_aspect(cor_aspect)
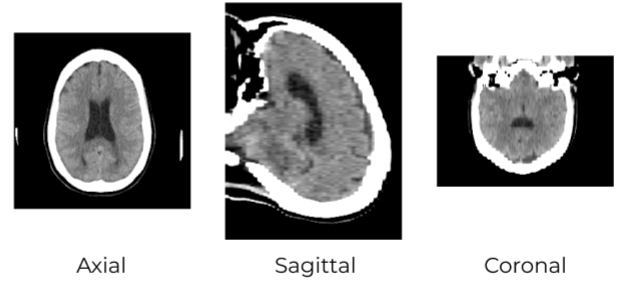
Multiplanar reformation or reconstruction (MPR) involves the process of converting data from an imaging modality acquired in a certain plane, usually axial, into another plane. It is most commonly performed with thin-slice data from volumetric CT in the axial plane, but it may be accomplished with scanning in any plane and whichever modality capable of cross-sectional imaging, including magnetic resonance imaging (MRI), PET and SPECT.
Windowing
level = dicom_file.WindowCenter
width = dicom_file.WindowWidth
# ...or set window/level manually to values you want
vmin = level - width/2
vmax = level + width/2
plt.imshow(hu_pixels, cmap='gray', vmin=vmin, vmax=vmax)
plt.show()


Brain windows are useful for evaluation of brain hemorrhage, fluid-filled structures including blood vessels and ventricles, and air-filled spaces.
Bone windows are useful for evaluation in the setting of trauma.
References
“What is the difference between slice thickness and slice increment?”
“Usefulness of hounsfield unit and density in the assessment and treatment of urinary stones”
“Multiplanar reformation (MPR)”. Dr Daniel J Bell and Dr Francis Fortin et al. radiopaedia.org
Follow me on twitter @theujjwal9