DICOM Standards
December 28, 2020
DICOM = Digital Imaging and Communications in Medicine
The DICOM standard includes a file format definition and a network communications protocol that uses TCP/IP to communicate between systems.
Abstract
The DICOM standard was conceptualized and implemented when CT (computed tomography) scans were developed. It takes special care not to lose information when translating medical images and other data to digital format. This is done to keep it as close to the original as possible.
The main objective of this new standard was to create an open platform for the communication of medical images and related data.
Introduction
The contents of the DICOM standard go far beyond a definition of an exchange format for medical image data.
DICOM defines:
- Data structures (formats) for medical images and related data
- Network-oriented services, e.g.:
- Image transmission
- Query of an image archive (PACS)
- Print (hardcopy)
- RIS - PACS - modality integration
- Formats for storage media exchange
- Requirements for conforming devices and programs
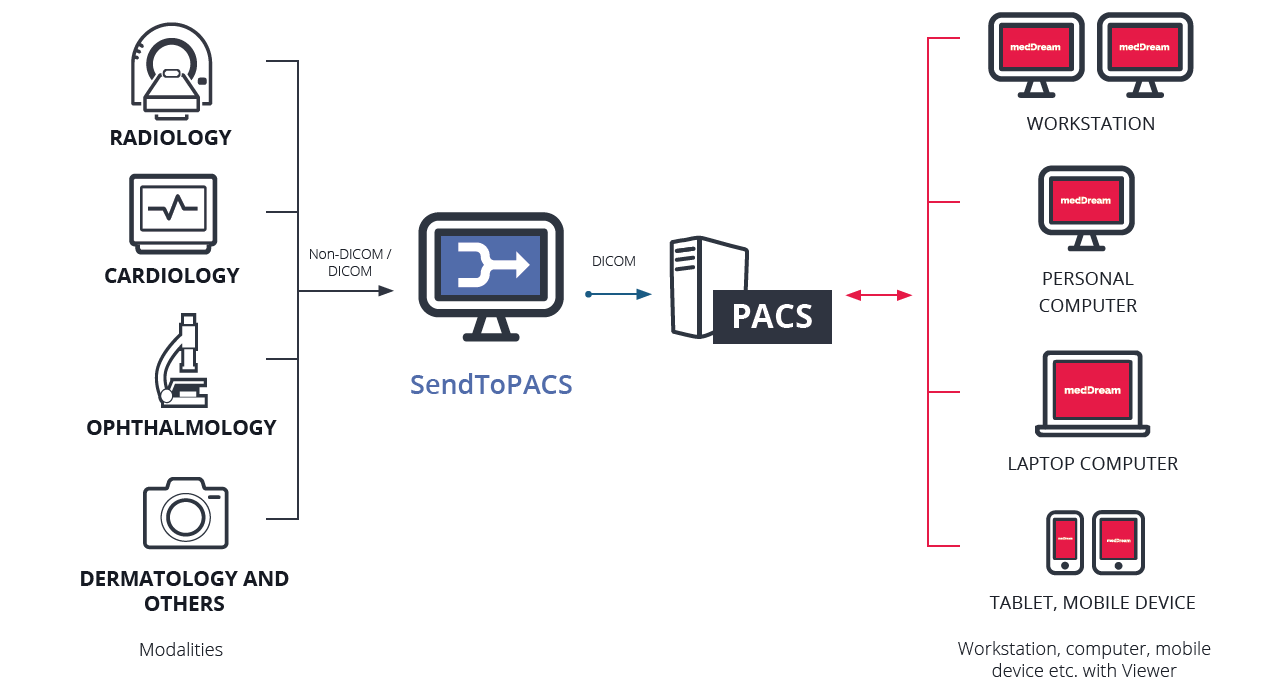
PACS (Picture Archiving and Communication System) provides economical storage of, and convenient access to, images from multiple modalities (source machine types).
RIS (Radiology Information System) is a computerized database used by radiology departments to store, manipulate, and distribute patient radiological data and imagery.
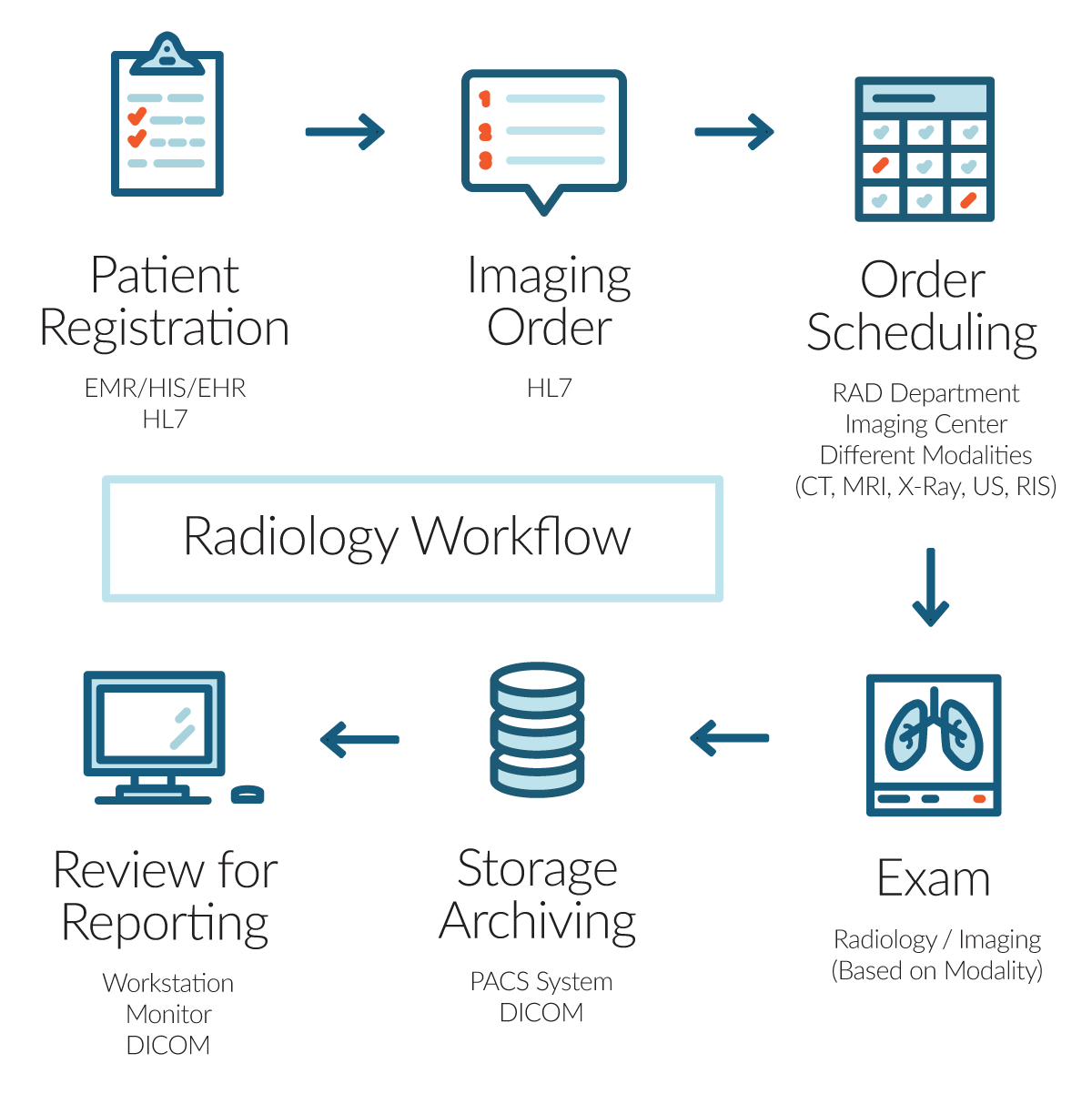
PACS-RIS integration improves the flow of images for radiologists. They communicate using a set of commands (called HL7) concerned with Admission/Discharge/Transfer (ADT) and Order/Entry.
DICOM Data Structures
DICOM consists of a list of image attributes which contain vast amounts of image and medical information:
- Patient information (name, sex, identification number)
- Modality and imaging procedure information (device parameters, calibration, radiation dose, contrast media)
- Image information (resolution, windowing)
DICOM goes to great lengths and defines the significance of each data element in a multitude of cases. It defines if an attribute is required, optional, or important for certain cases. But this feature comes at a cost:
- Image objects are frequently incomplete: There is inconsistency in filling all the fields with data. Some fields in image objects are often left blank and some are filled with incorrect data.
- Another problem occurs when displaying an image on a device made by a different manufacturer, because different imaging equipment uses different amplitude ranges and the same number of allocated bits. In that case, images can be displayed as underexposed or overexposed with poor contrast, so those parameters should be adjusted manually.
DICOM Network Services
This service is based on the client-server concept. DICOM applications establish connections to exchange information. In addition to image transmission, there are other features:
- Image Archive Service: Search images in a PACS archive by certain criteria (patient, time of creation of the images, modality etc.) and selectively download images from this archive
- Print Service: Gives access to cameras and printers over a network
- Modality Worklist Service: Download updated information regarding patients using the above-described PACS-RIS system
Patient Model
Queries to image archives (PACS) are made in 4 levels of DICOM hierarchy:
Patient has studies, studies have series which are scans and scans may have multiple instances or images (which are slices in CT scans).
Patient Level
- How many studies are there for this patient
- How many Series are there for this patient (in all studies)
- How many Instances (images) are there for this patient (in all series)
Study Level (Patient and Study roots)
- How many Series are there in this study
- How many Instances (images) are there in this study (in all series)
Series Level
- How many Instances (images) are there in this series
Instance Level
Table is sourced from here
| Attribute Name | Tag | Attribute Description |
|---|---|---|
| Number of Patient Related Studies | (0020,1200) | The number of studies that match the Patient level Query/Retrieve search criteria |
| Number of Patient Related Series | (0020,1202) | The number of series that match the Patient level Query/Retrieve search criteria |
| Number of Patient Related Instances | (0020,1204) | The number of composite object instances that match the Patient level Query/Retrieve search criteria |
| Number of Study Related Series | (0020,1206) | The number of series that match the Study level Query/Retrieve search criteria |
| Number of Study Related Instances | (0020,1208) | The number of composite object instances that match the Study level Query/Retrieve search criteria |
| Number of Series Related Instances | (0020,1209) | The number of composite object instances in a Series that match the Series level Query/Retrieve search criteria |
| SOP Classes in Study | (0008,0062) | The SOP Classes contained in the Study. |
Media Exchange
DICOM defines application profiles which defines how media is exchanged:
- Encoding formats and compression schemes used (e. g. only uncompressed or loss-less JPEG)
- Storage medium used
- Images from which modalities may be present on the medium (X-Ray Angiography images, etc)
DICOM directory: each dicom medium contains this directory which contains information (patient name, modality, unique identifiers etc.) for all images which are captured on the medium.
Device Conformance
Each DICOM supporting device must also specify which DICOM services and options are supported, which extensions and peculiarities have been implemented by the vendor, and how the device communicates with other DICOM systems.
DICOM File Format
The DICOM standard is divided in 2 parts:
A DICOM file consists of a header and image data sets.
 Packet structure of dicom file.
Packet structure of dicom file.
Preamble is used to access the images and other data in DICOM file.
Prefix contains the string “DICM” as uppercase characters.
Data Set is the representation of real world information.
Data Elements. There are 5 types of Data elements:
- Type 1 Required Data elements,
- Type 1C Conditional Data Elements,
- Type 2 Required Data Elements,
- Type 2C Conditional Data Elements
- Type 3 optional Data Elements.
Clinical Terms
Some clinical terms to be aware of!
Slice Thickness
Slice thickness refers to the (often axial) resolution of the scan.
Patient Position (0018, 5100) will tell you if the patient was scanned head-first supine, feet-first prone, head-first prone, etc. Instance Number (0020, 0013), also commonly known as slice number, contains no information about spatial location and isn't even guaranteed to be unique. Slice Location (0020, 1041) is useful, if it exists, but you can't count on it always existing because it's a Type 3 (optional) attribute. To have a robust solution, you need to use Image Position Patient (0020, 0032) together with Image Orientation Patient (0020, 0037) and Patient Position (0018, 5100) to properly order the slices in space. Image Position Patient gives you a vector from the origin to the center of the first transmitted pixel of the image. Image Orientation Patient gives you vectors for the orientation of the rows and columns of the image in space. Patient Position tells you how the patient was placed on the table relative to the coordinate system.
Hu Scaling (Hounsfield scale)
HU is a quantitative scale for describing radiodensity. HU’s is standardized across all CT scans regardless of the scanner detector.
\[HU = 1000 * \frac{\mu - \mu_{water}}{\mu_{water} - \mu_{air}}\]Where \(\mu\) is linear attenuation coefficient.
Here’s a quick list of a few useful ones, sourced from Wikipedia.
| Substance | HU | |
|---|---|---|
| Air | −1000 | |
| Fat | −120 to −90 | |
| Soft tissue on contrast CT | +100 to +300 | |
| Bone | Cancellous | +300 to +400 |
| Cortical | +1800 to +1900 | |
Windowing
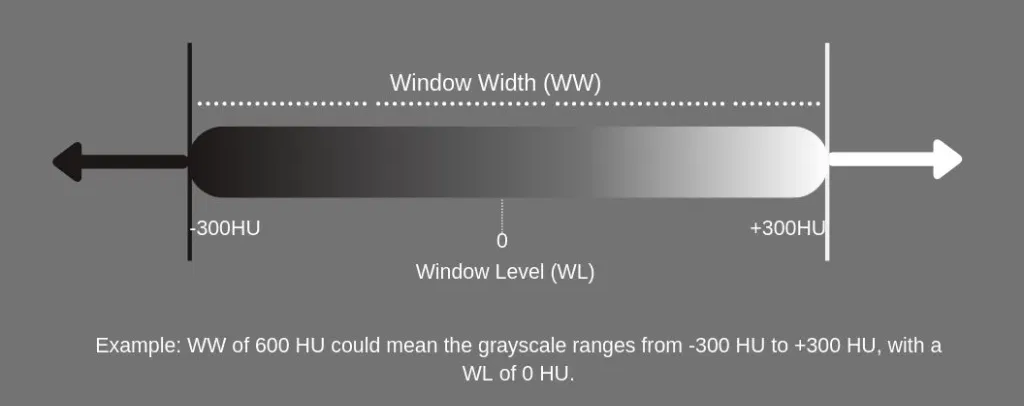
Since it is difficult to recognize 4000 shades of gray easily, we use windowing. It limits the number of Hounsfield units that are displayed.
For example, if we want to examine the soft tissue in one CT scan we can use a window level of 40 and a window Width of 80 this will cover 40 units below and above the window level and the tissues with CT numbers outside this range will appear either black or white. A narrow range provides a higher contrast.
The window width is the range of the grayscale that can be displayed. The center of grayscale range is referred to as the window level.
You can go through this blog to learn more about windowing.
Walkthrough Ohif Viewer
This section contains steps on how to use ohif viewer for viewing dicom file based medical records (studies, series, instances).
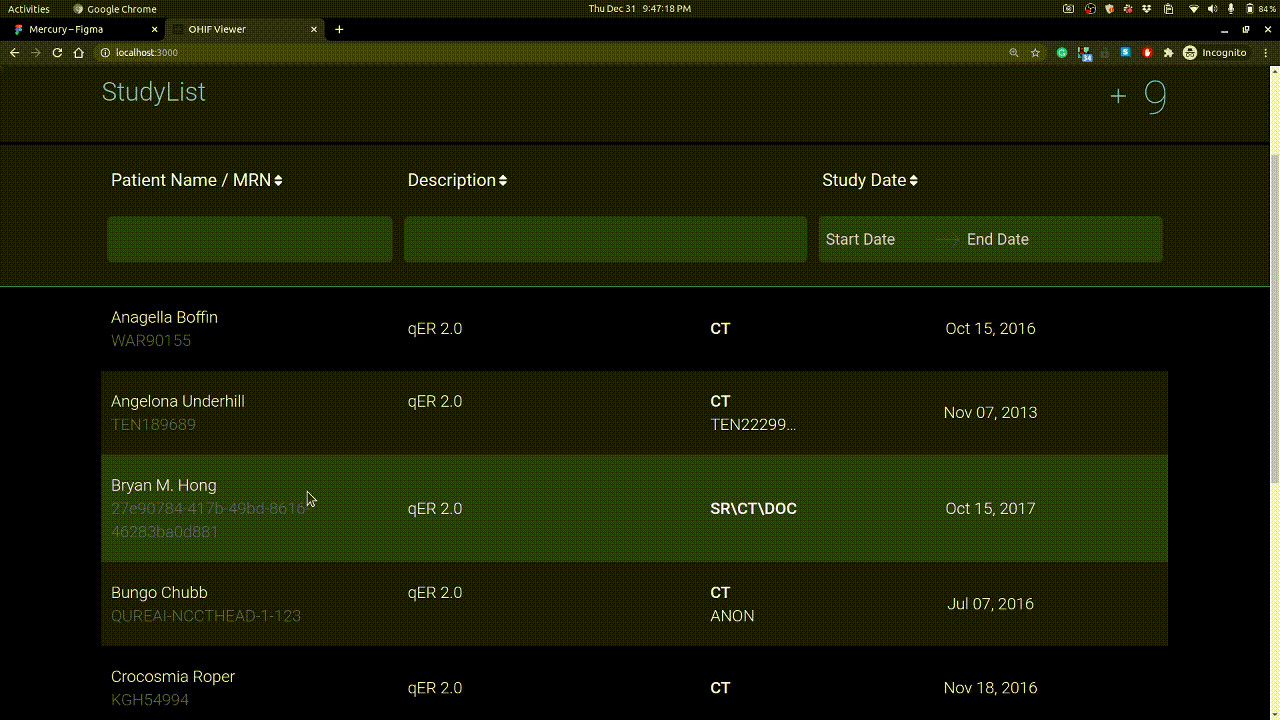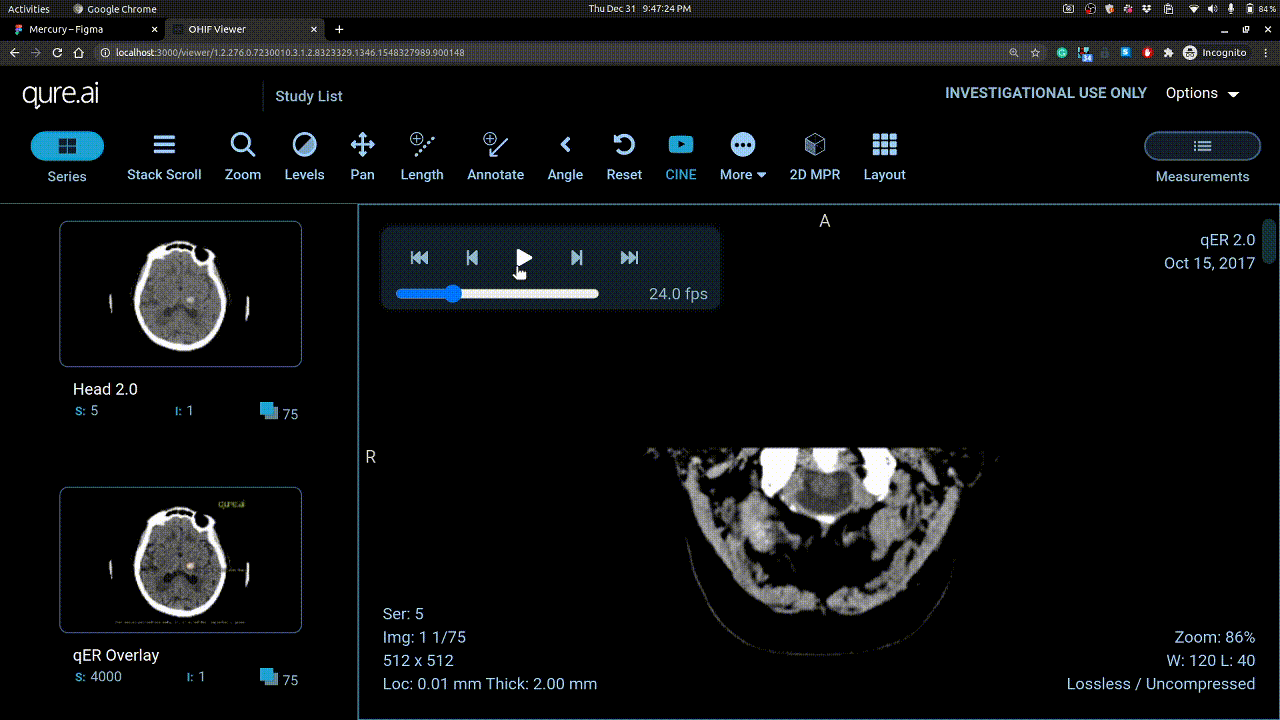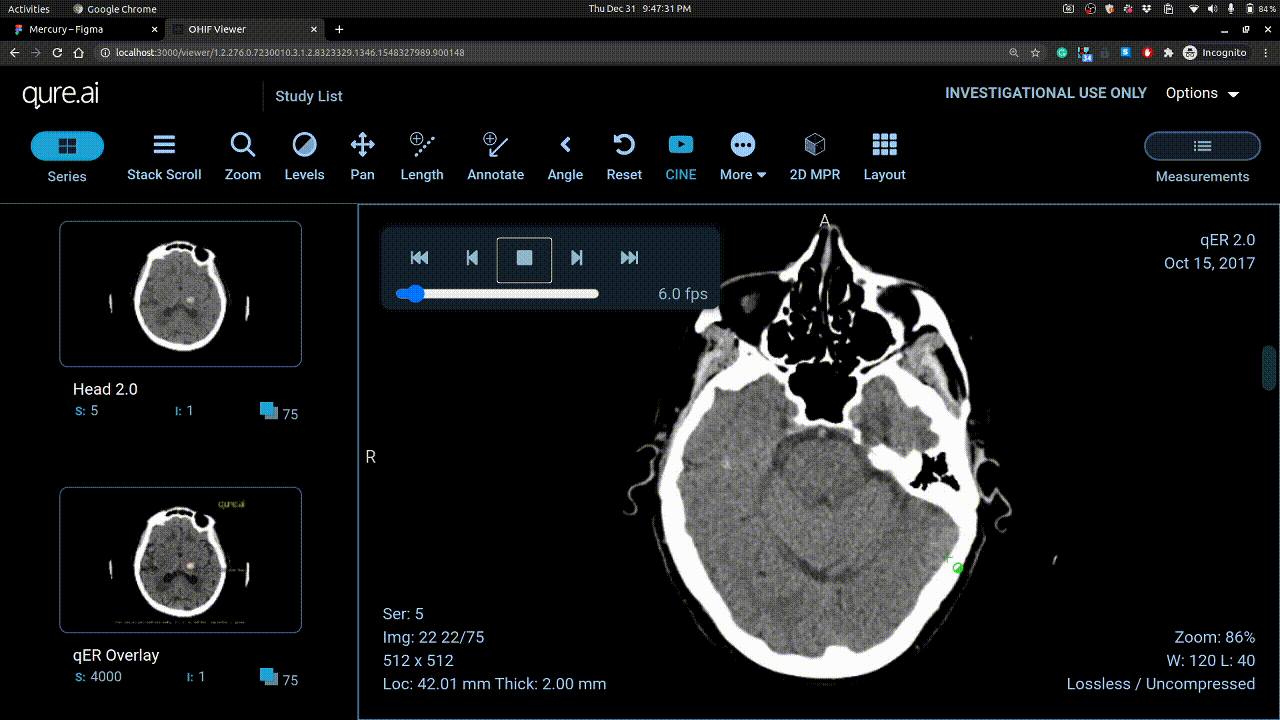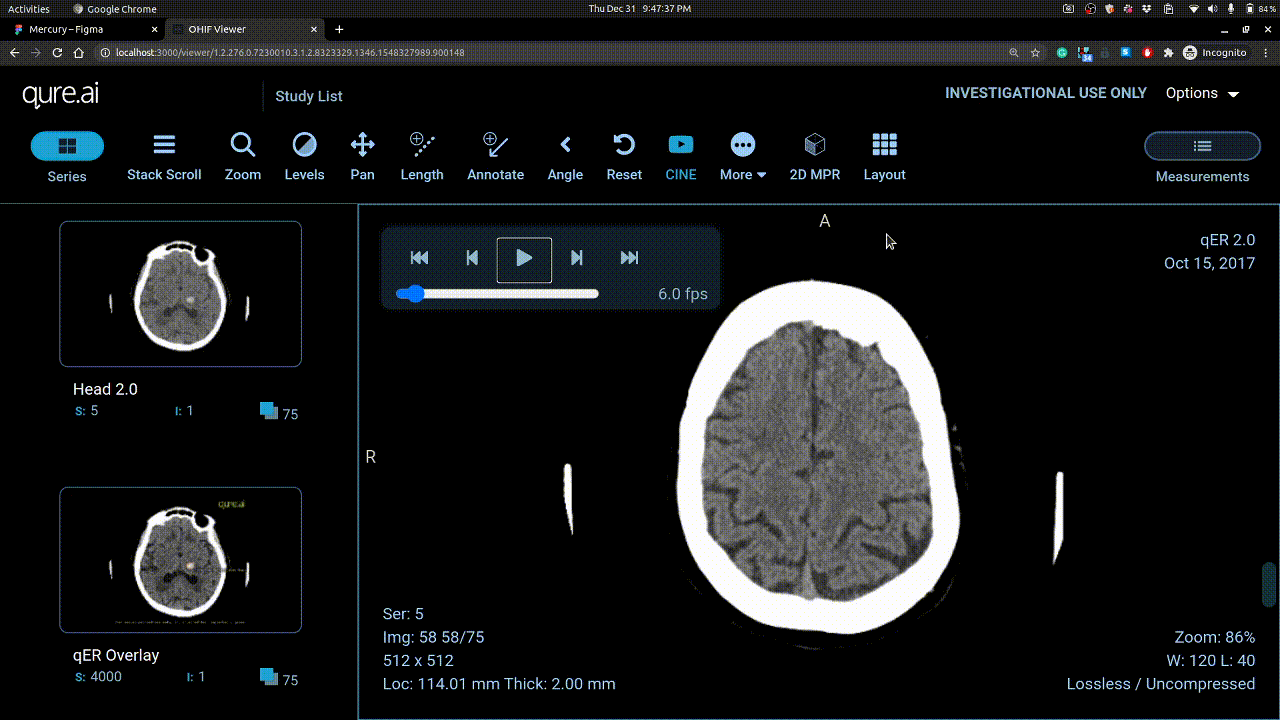
Homepage
Here you see the studies you have uploaded. Each study have series and these series have instances / images.
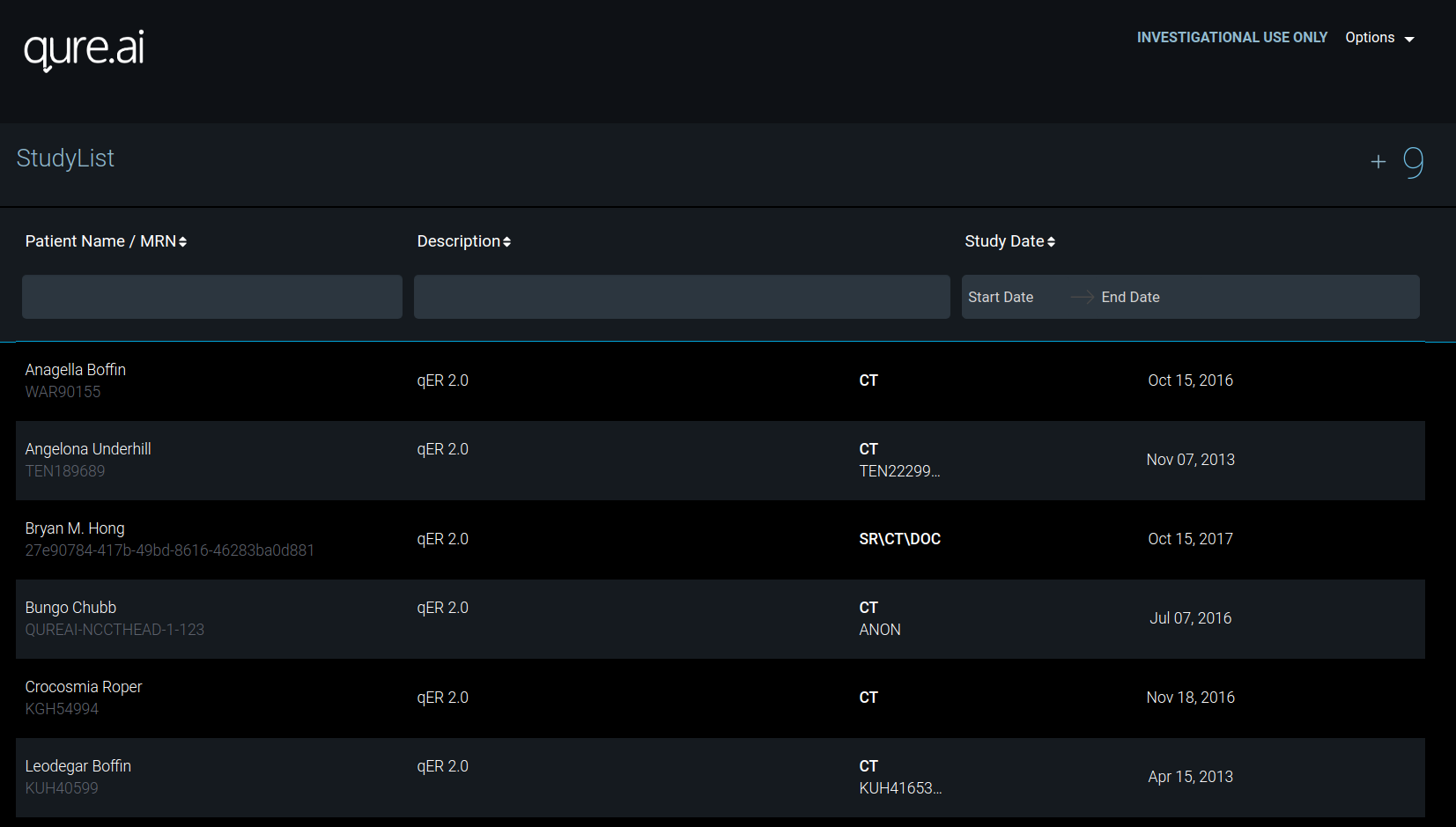
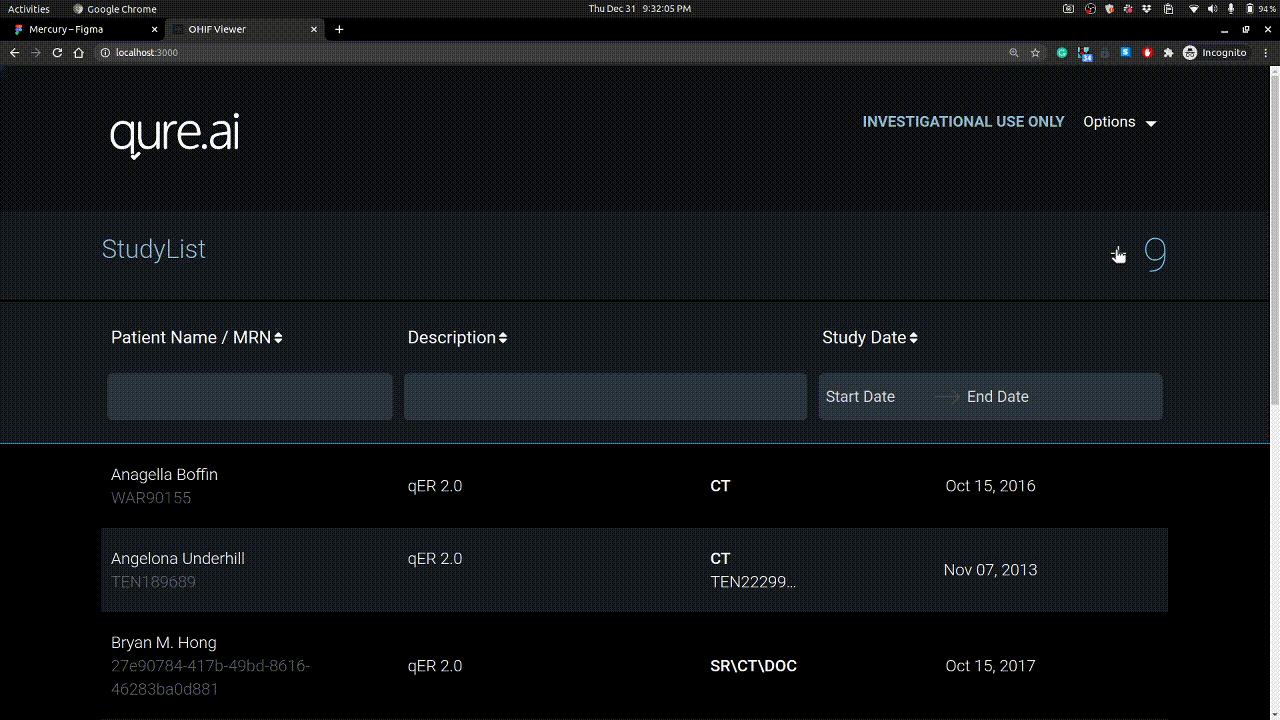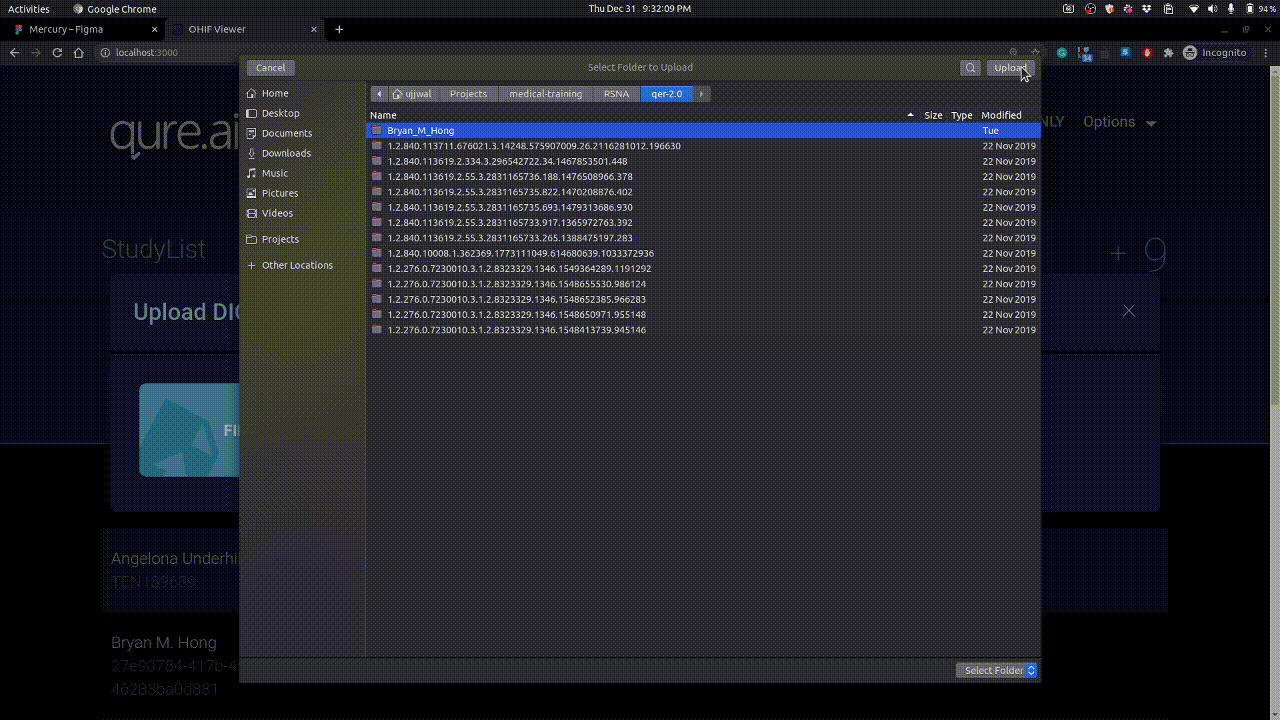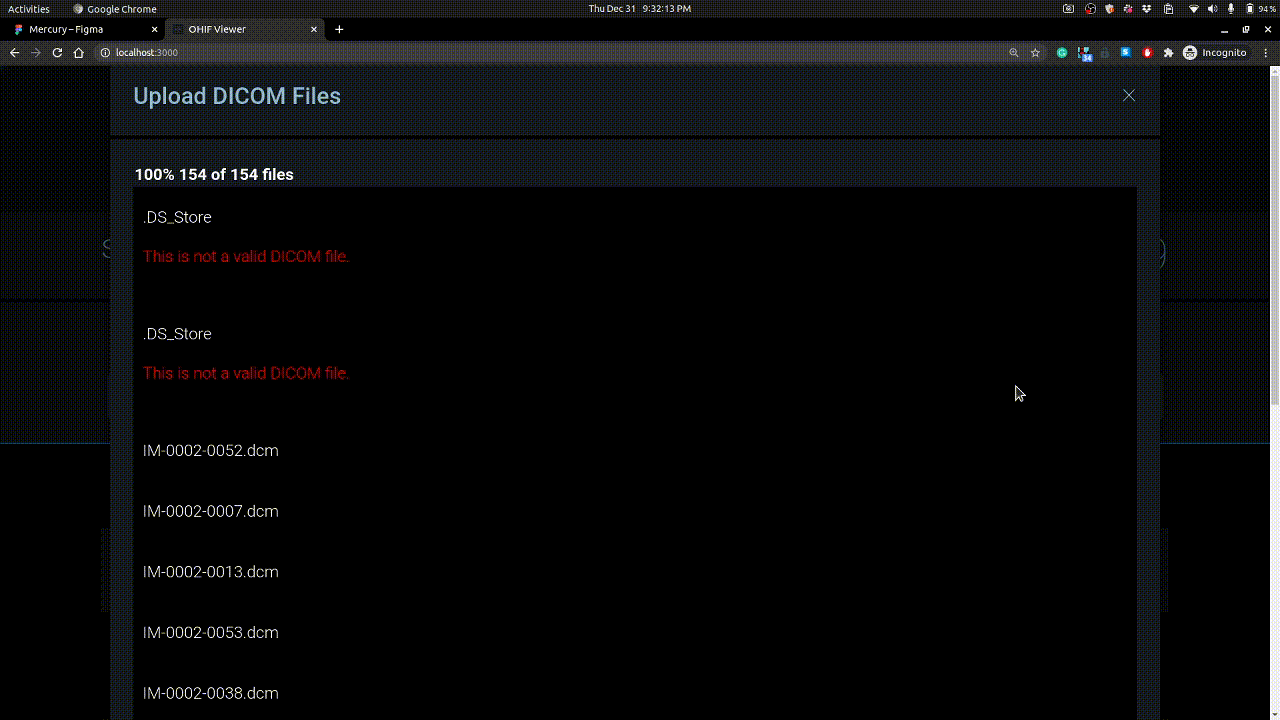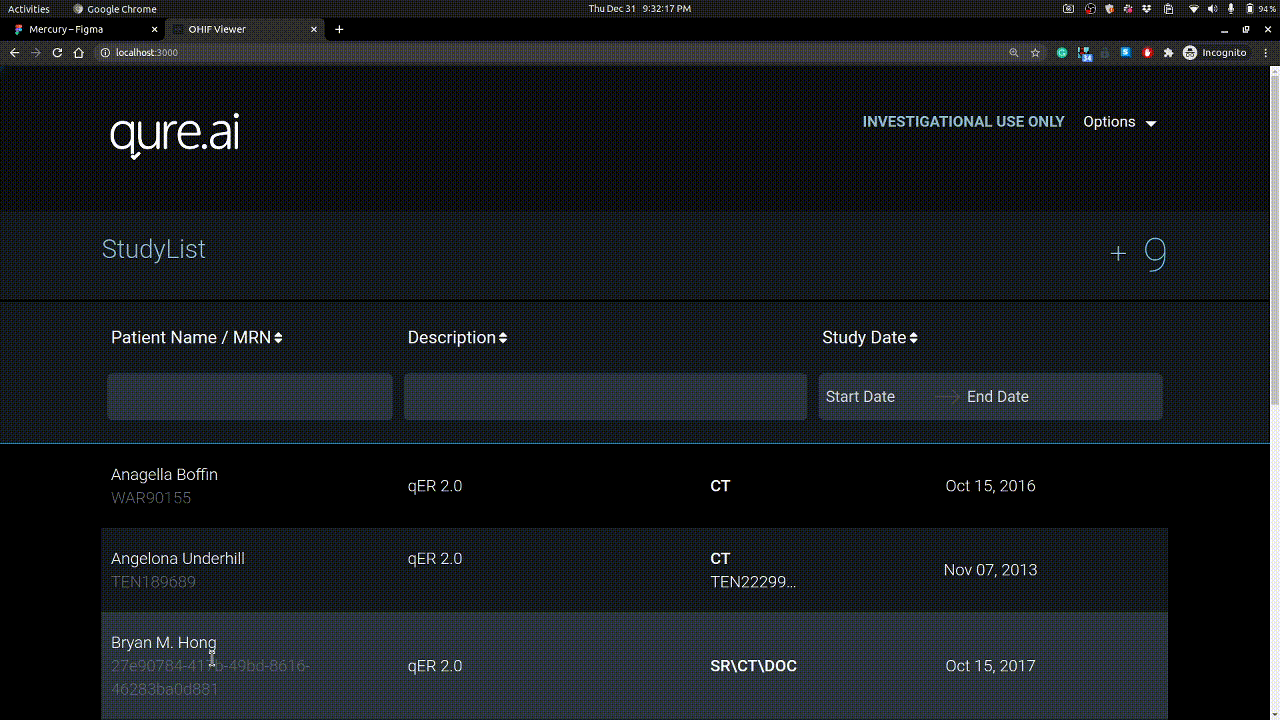
Studies can be uploaded using + button on right. Example is shown below.
Details on Scan View
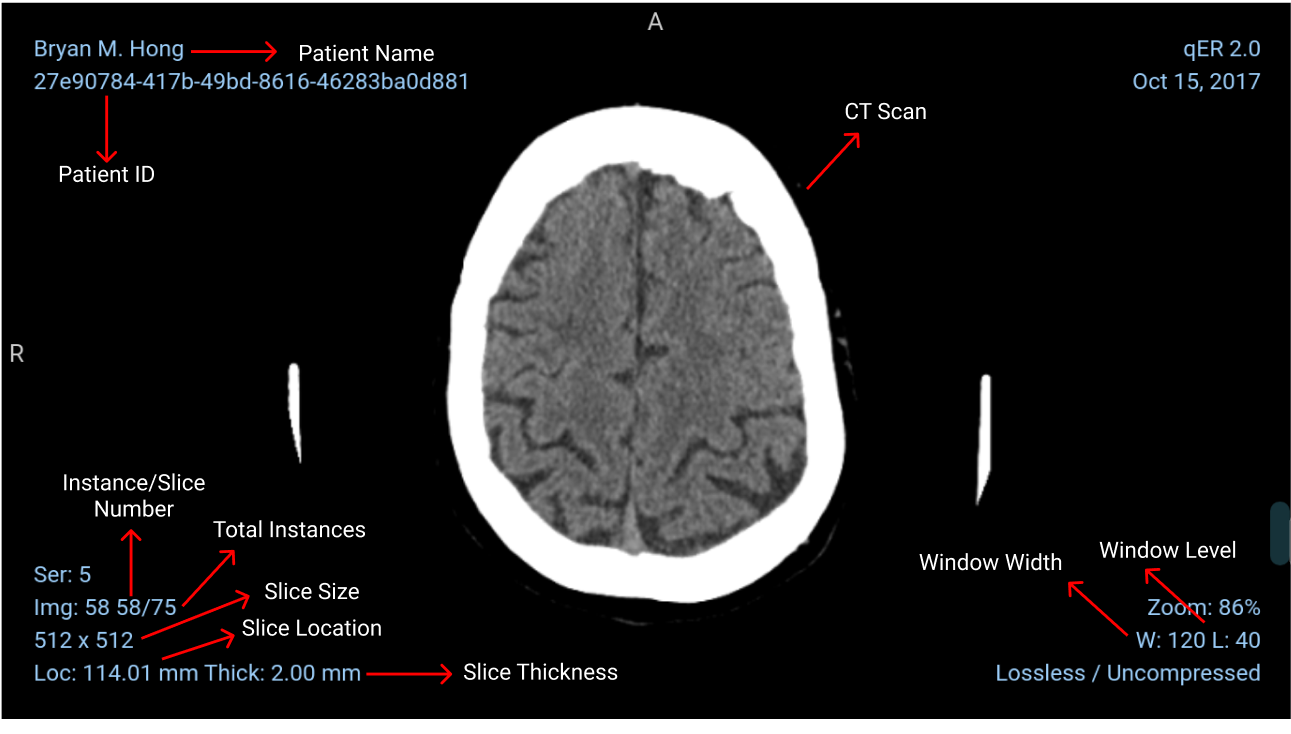
Calibrating Windowing
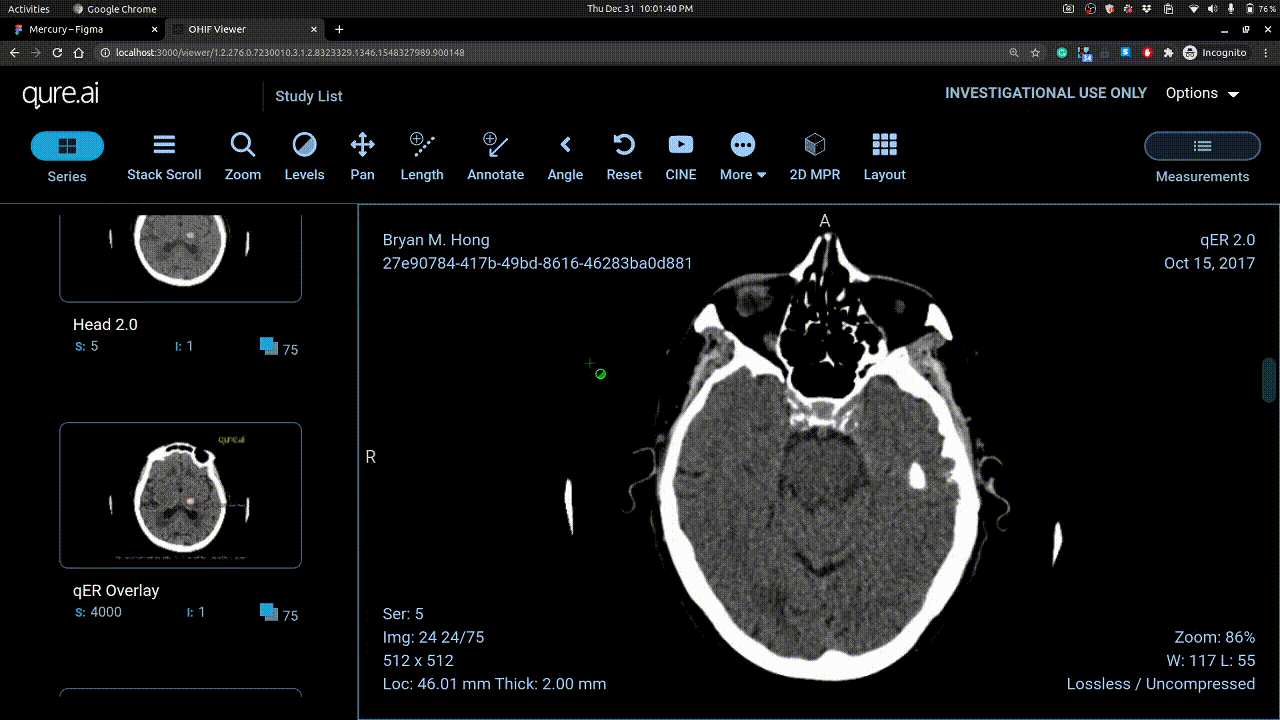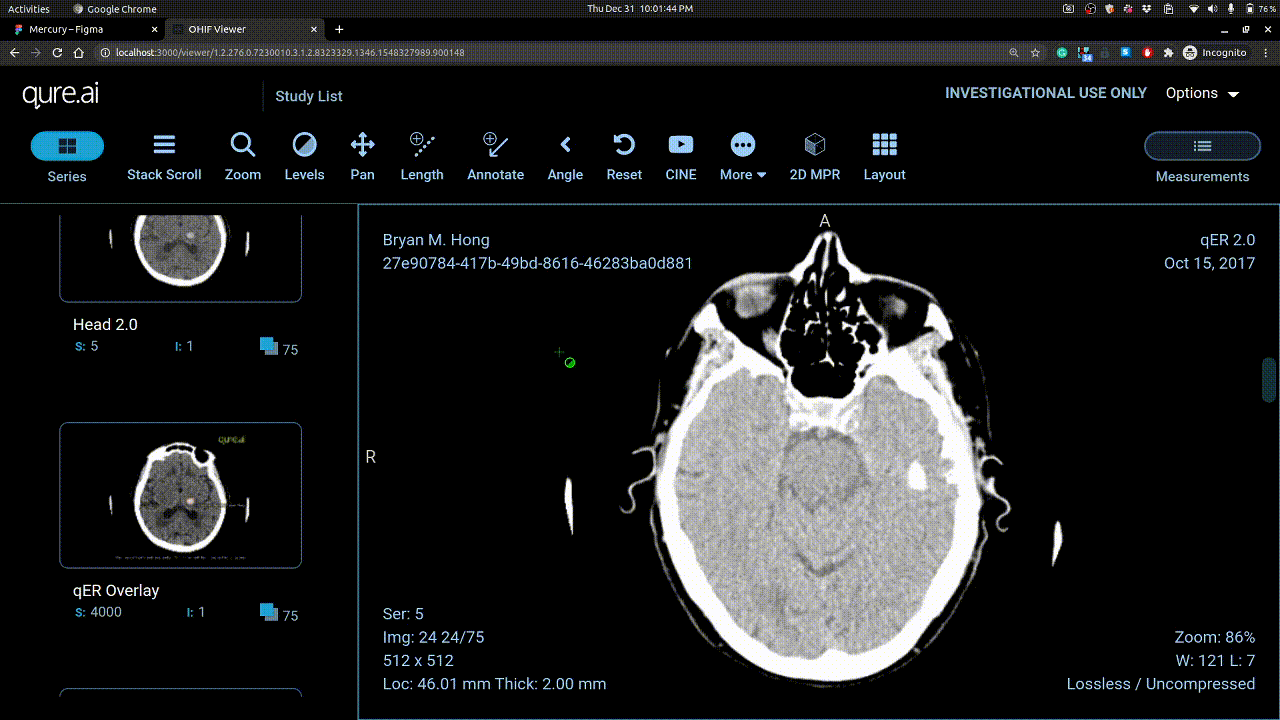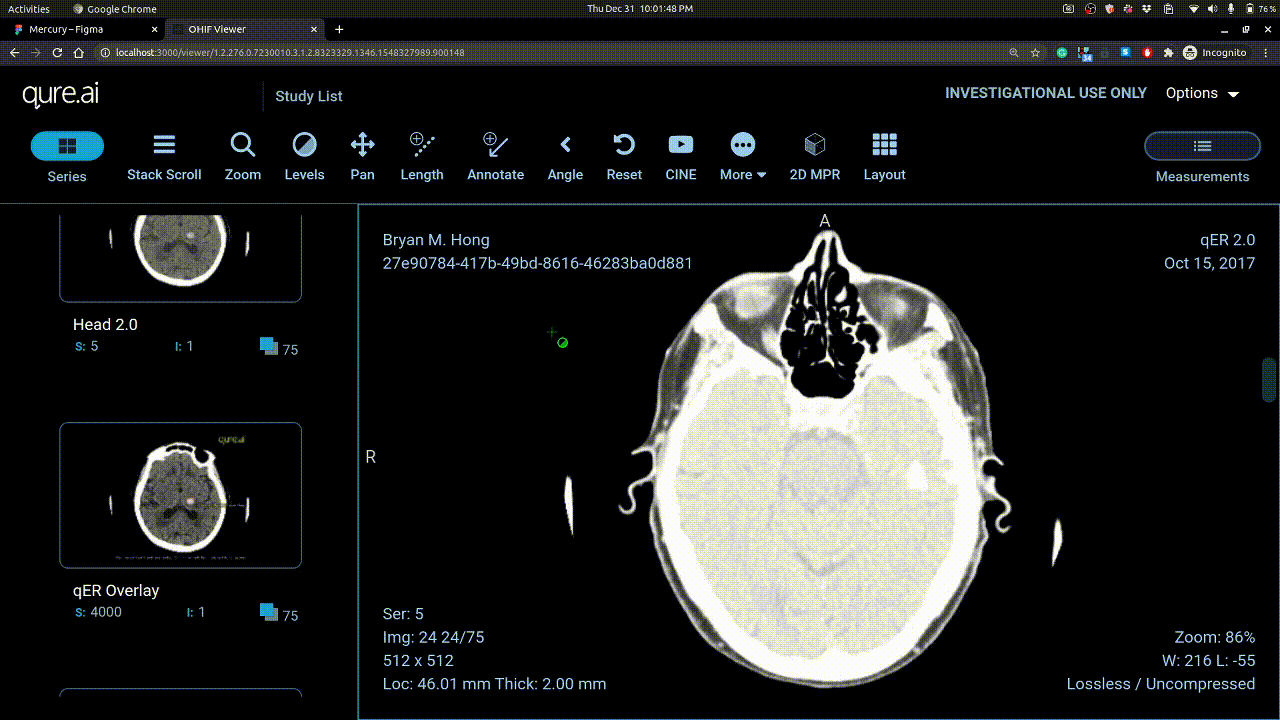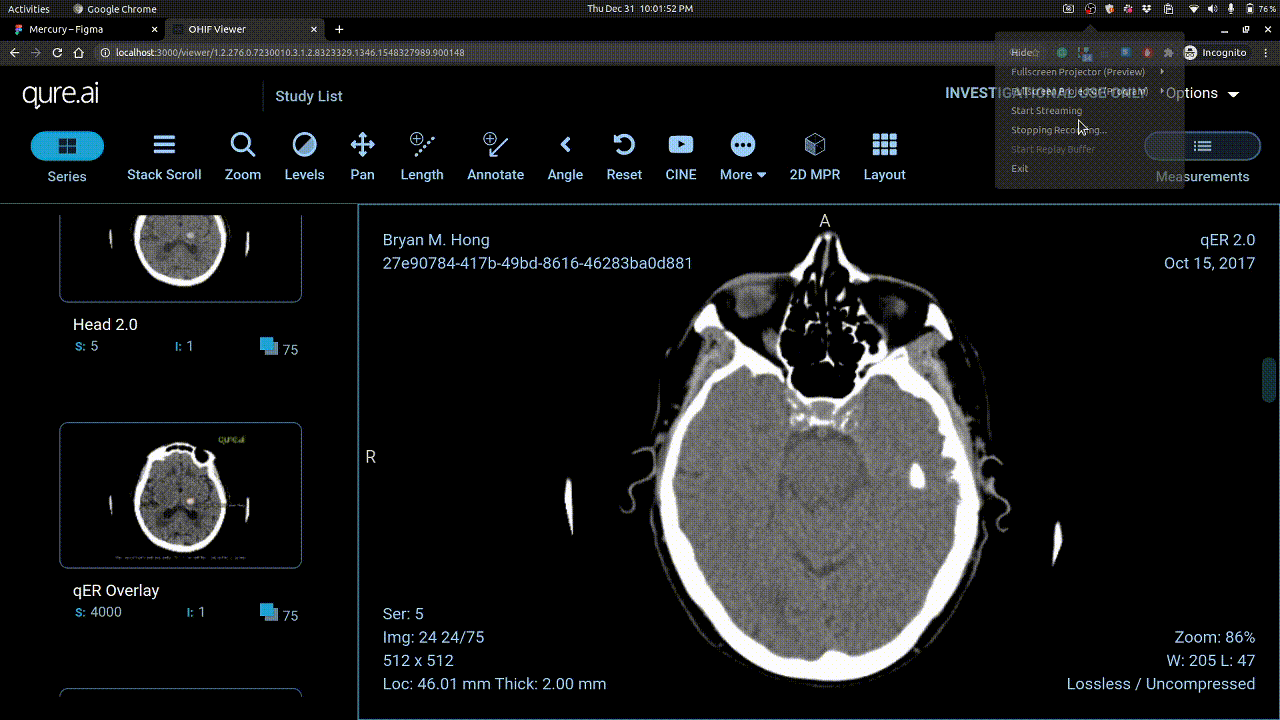
Vertical mouse movement changes window level and horizontal movement changes window width.
Visualization of slices
2D Multiplanar Reconstruction (MPR)
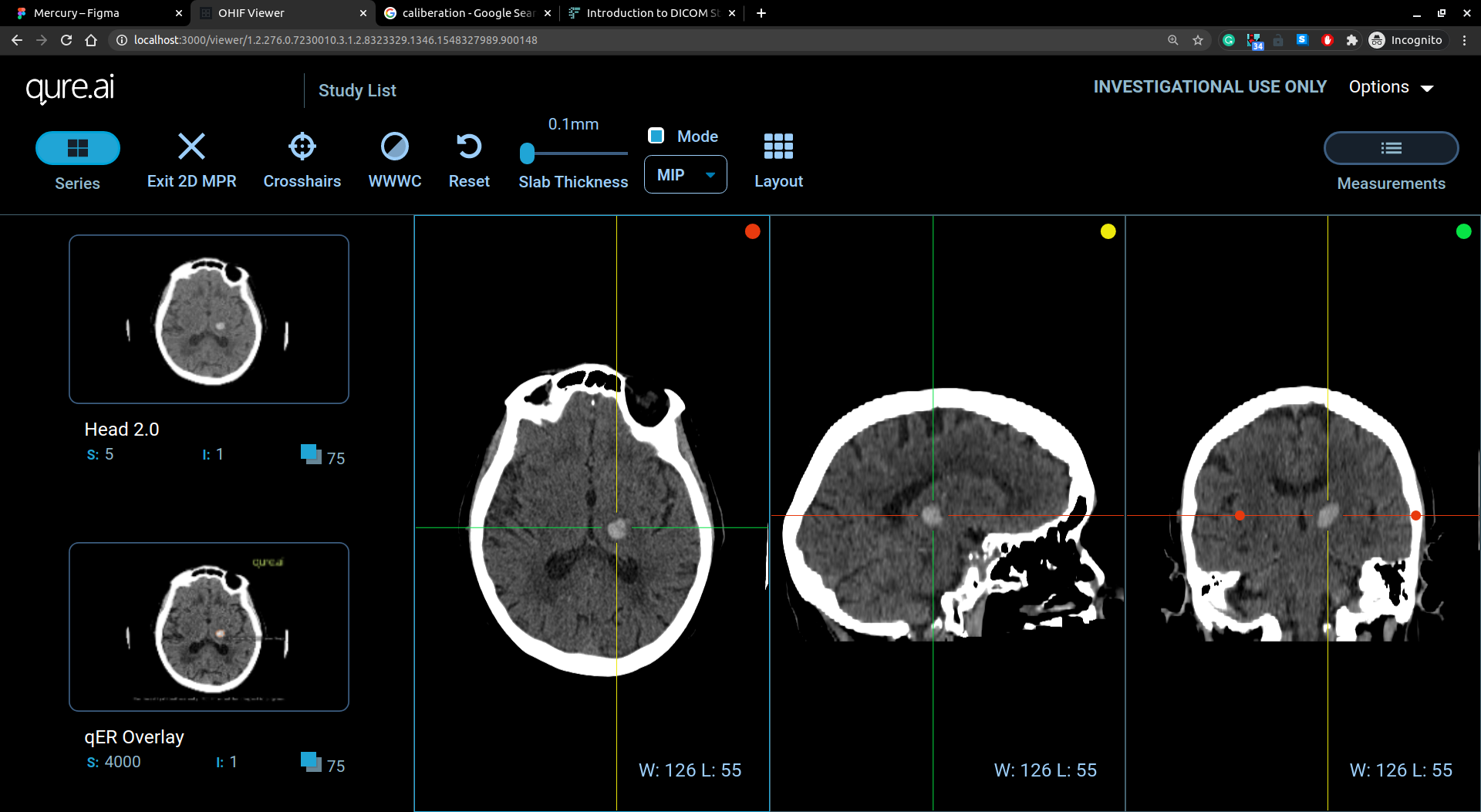
The MPR tool provided within the Viewer can be used to reconstruct images in orthogonal planes (coronal, sagittal, axial or oblique, depending on what the base image plane is). This can help to create a visualization of the anatomy which was not possible using base images alone.
Report view
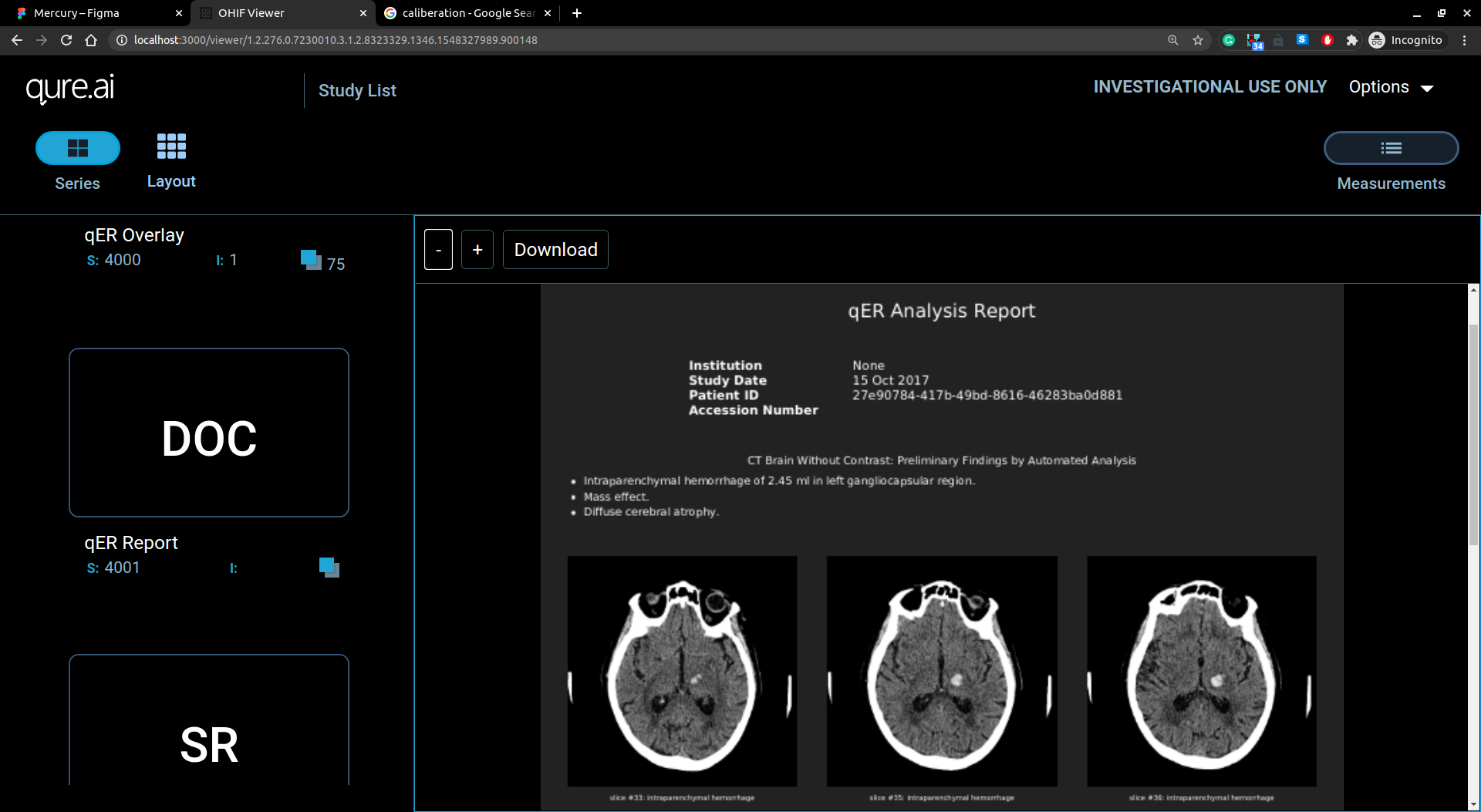
Report view has report on doc/pdf format displayed in viewer.
References
“Picture archiving and communication system”. medico-eng.com
“The case for RIS/PACS integration”. PUBMED NCBI
“Counting Studies, Series and Instances”. medicalconnections.co.uk
Follow me on twitter @theujjwal9